Displaying Code in LaTeX
gioby of Bioinfo Blog! (an interesting read by the way) left a comment asking about displaying code in LaTeX documents. I’ve sort of been cludging around using \hspace‘s and \textcolor but I’ve always meant to figure out the right way to do things so this seemed like a good chance to figure out how to do it right.
LaTeX tends to ignore white space. This is good when you’re writing papers but not so good when you’re trying to show code where white space is an essential part (e.g. Python). Luckily there’s a builtin verbatim environment in LaTeX that is equivalent to html’s <pre>. So something like the following should preserve white space.
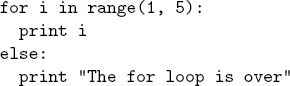
\begin{verbatim}
for i in range(1, 5):
print i
else:
print "The for loop is over"
\end{verbatim}Unfortunately, you can’t use any normal LaTeX commands inside verbatim (since they’re displayed verbatim). But luckily there a handy package called fancyvrb that fixes this (the color package is also useful for adding colors). For example, if you wanted to highlight “for” in the above code, you can use the Verbatim (note the capital V) environment from fancyvrb:
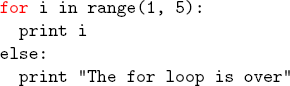
\newcommand\codeHighlight[1]{\textcolor[rgb]{1,0,0}{\textbf{#1}}}
\begin{Verbatim}[commandchars=\\\{\}]
\codeHighlight{for} i in range(1, 5):
print i
else:
print "The for loop is over"
\end{Verbatim}
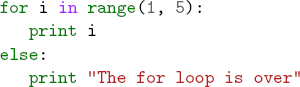
If you really want to get fancy, the Pygments package in Python will output syntax highlighted latex code with a command like: pygmentize -f latex -O full test.py >py.tex The LaTeX it outputs is a bit hard to read but it’s not too bad (it helped me figure out the fancyvrb package) and it does make nice syntax highlighted output.
Here’s an example LaTeX file with the three examples above and the pdf it generates if you’re curious.
